Genome Sequence is Deciphered for Endangered Himalayan Medicinal Herb Picrorhiza Kurrooa (Kutki)
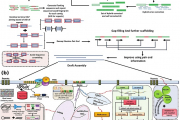
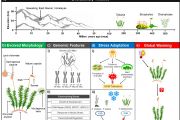
Team CSIR-IHBT published the draft genome of Picrorhiza kurrooa (Kutki) in an international journal Scientific Reports, a Springer Nature publishing group (https://www.nature.com/articles/s41598-021-93495-z). Earlier, the group had also published the transcriptome of the species in the year 2012. The present study brings the first assembled draft genome of P. kurrooa. It has a large size (2n=3.4 GB) and is much more complex (>75% complex repeats). The entire work was conducted in-house at CSIR-IHBT, Palampur. From a single institution, it is one of the biggest draft genome of a plant species being reported from India and also the first eukaryotic Himalayan-medicinal-herb genome. The assembled draft genome of P. kurrooa offers a valuable resource and reference information to understand biology of this important endangered Himalayan medicinal plant. There is a great challenge in solving the genome of eukaryotes. Usually, the sequences are repeated because of which it becomes very difficult to assemble the genome. Herein, we adopted a special computational method to assemble the genome in spite of high repeats.
Decoding the sequence of DNA that constitutes the genome is imperative to biological research. The sequence of nucleic acids in DNA eventually contains the information for heredity and synthesis of biomolecules. Integrating genome sequencing into medicinal plant research is perceivable as it could shed light on biosynthesis of medicinally active compounds as well as play a major role in understanding of complex molecular mechanisms to aid in improvement and conservation of medicinal plants. Considering high medicinal value and market demand of plant derived natural products, researchers are now focusing on development of genomic resources for medicinal plants. The genomes of medicinal plant species including Catharanthus roseus, Salvia miltiorrhiza and Camptotheca acuminata have been sequenced. Genome sequence information has allowed better understanding of poorly understood areas of medicinal plants including secondary metabolite biosynthesis as well as adaptation to various environmental cues.
P. kurrooa is an endangered medicinal herb that grows in Himalayan regions of India, China, Pakistan, Bhutan and Nepal and is distributed across the Himalayan region at an altitude between 3000–5000 m above mean sea level. The medicinal properties of P. kurrooa are attributed to picrosides present in leaf, rhizome, and root of the plant. It is widely harvested from natural habitat for use in traditional ayurvedic system of medicine. The species is used in a number of commercially available drug formulations like livocare, livomap, livplus and katuki for the treatment of liver ailments. Recent increased global demand for herbal medicines has posed additional pressure on naturally occurring populations of P. kurrooa. A high demand in the market coupled with unorganized collection from the wild sources together with lack of cultivation have threatened the species to near extinction, making it listed as endangered species. Keeping in view the medicinal and economical importance of P. kurrooa, and its endangered status, it was essential to sequence the whole genome that is expected to aid in improvement, better molecular interventions, and conservation of the species.